Ligands, Linkers & Tools Available from key organics
The concept of Targeted Protein Degradation (TPD) emerged from the laboratory of Craig Crews at Yale University following a seminal 2001 publication. In a very simplified form, this can be viewed as using a carefully designed “three component” compound (E3 ligase binder, linker, protein-of-interest binder) to harness the cells endogenous waste disposal machinery (the Ubiquitin-Proteasome System, UPS) which results in the targeting of specific proteins for degradation. The 5-step process of protein / ligase binding, polyubiquitinylation and ultimately proteasomal degradation is summarised schematically in Figure 1.
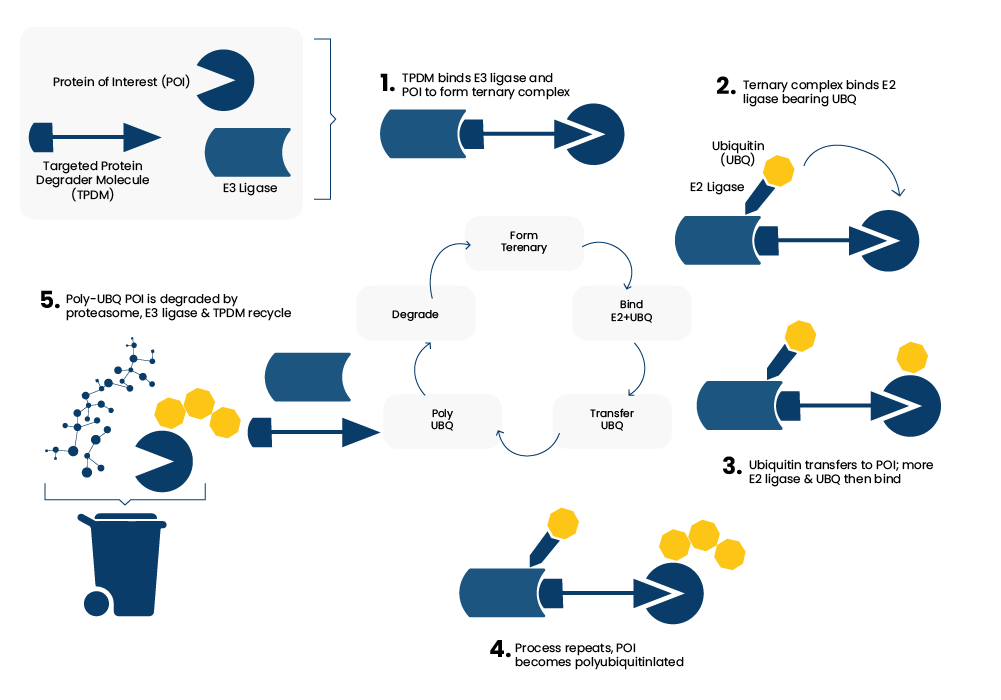
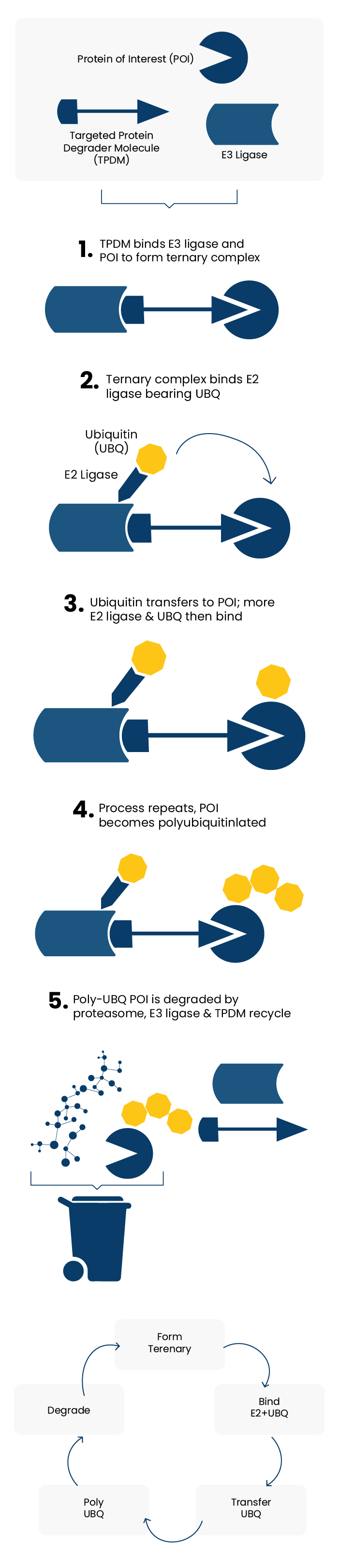
Figure 1 : The Targeted Protein Degradation Concept
Although targeted protein degradation molecules (TPDM) occupy significantly different chemical space compared to traditional ‘Lipinski-compliant’ small molecule drugs, they have been noted as an attractive concept when applied to the development of therapeutic agents because the whole protein is targeted, not just an active site. This widens the range of targets beyond pure enzymes, and also renders the approach less resistant to mutational bypass which is often seen for enzyme inhibitors. This is particularly relevant for therapeutic areas where such resistance can be a serious issue, including oncology and anti-infectives. In principle the targeted degradation process is also catalytic, which allows the active compound to function in a sub-stoichiometric fashion and be very atom efficient.
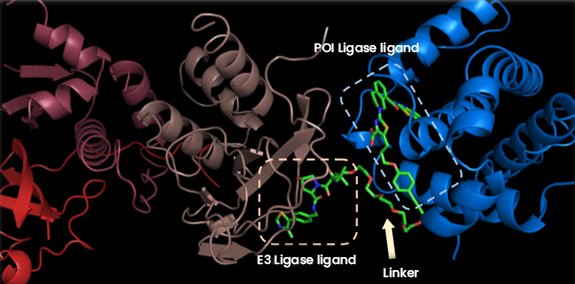
Figure 2 : The ElonginB:ElonginC:VHL/TPDM/Bcl-xL complex (RCSB PDB code 6ZHC)
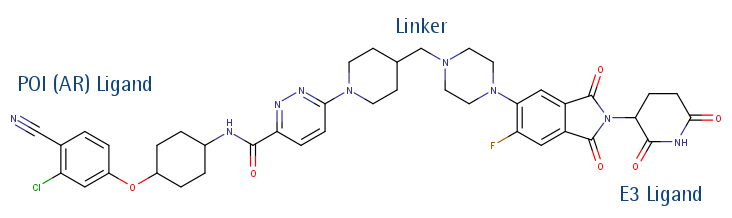
Since Crew’s original publication, literature interest in the area has mushroomed, with hundreds of publications a year. The increasing interest in TPDMs from the chemical biology and drug discovery community has arisen for two important reasons. Firstly the compounds can be utilised as tools for studying important biological processes, where they can be viewed as a type of ‘chemical knockdown’ akin to RNAi methodologies. Secondly the strategy offers significant potential to provide a brand new class of drugs for therapeutic use, and the first drugs using a targeted protein degradation approach such as ARV-110 are now entering mid-stage clinical trials (Figure 3).
ARV-110 (Arvinas)
Targeted Protein Degradation Citations (PubMed)
Figure 3 : Targeted protein degradation citations from 2001–20; ARV-110, the first clinical candidate
The field has attracted considerable interest and investment from the venture community, and multiple companies have emerged in recent years utilising TPD or related approaches as part of or all of their discovery pipelines across different therapeutic areas. An increasing number of compounds are in the late stages of optimisation, with several also approaching or in human clinical trials.
It is noteworthy that the ‘supersized’ physicochemical characteristics of TPD molecules have attracted attention as a potential limitation to the field for oral drugs. Consequently researchers have undertaken in silico evaluation of the area and work is ongoing in order to establish the optimal strategies to mitigate this and enable suitable PK properties for systemic delivery or CNS-accessible protein degradation agents.
From a mechanistic perspective it has also been recognised that there are subtle variations possible within the TPD arena, from the concept of simple molecular glues to detailed evaluation of strategies for more efficient E3 ligand identification, linker design, protein affinity aspects as well as widening the range of proteins that can be targeted effectively. In recent years the original approach has also expanded beyond targeting proteins alone to facilitating degradative strategies towards a broader range of biomolecules, including the lysosome (Lysosome-Targeting Chimeras, LYTAC) and RNA (Ribonuclease-Targeting Chimeras RIBOTAC).
Key Organics has a range of off-the-shelf products available for workers in the TPD field. We provide support for academic, biotech and pharma customers in this area through our growing range of catalogue ligands, linkers and tool compounds. Our bespoke fragment collection provides a rich resource for discovery of new ligands for both E3 ligases and target proteins of interest. We routinely undertake synthesis of bespoke or non-commercial compound(s) on either a custom fee-for-service (FFS) or contract (FTE) basis to support Hit Identification, Hit-to-Lead and Lead Optimisation activities for your discovery program.
Selected E3 ligase binders and functionalised building blocks, available from Key Organics
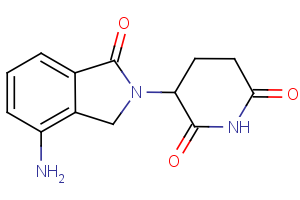
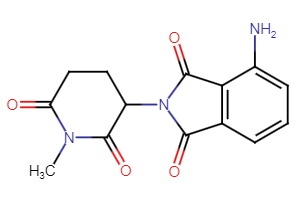
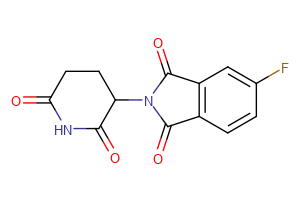
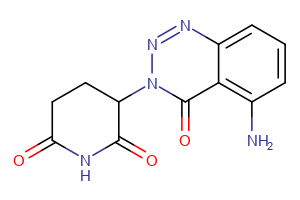
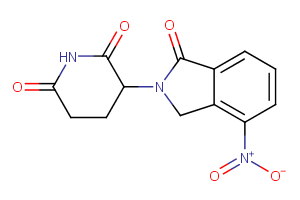
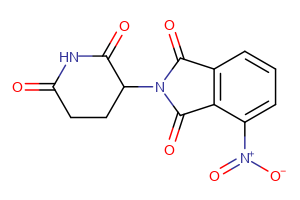
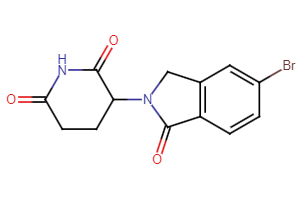
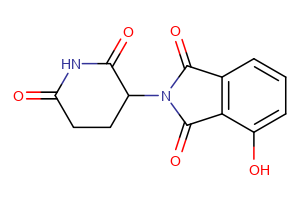
CRBN Ligands
VHL Ligands
Selected linker groups for connecting E3 and POI binding motifs, available from Key Organics
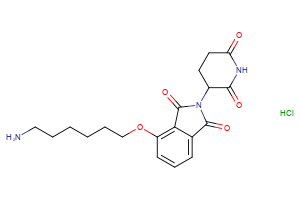
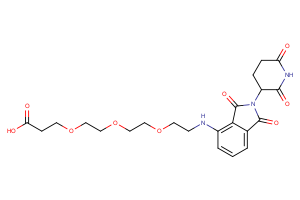
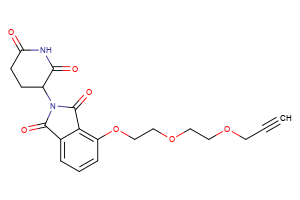
Selected “hemi-TPDM” building blocks (E3 binder + linker), available from Key Organics
E3 ligand with pre-appended linker
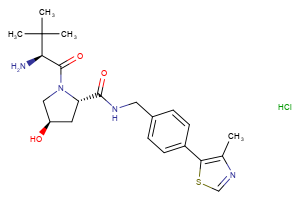
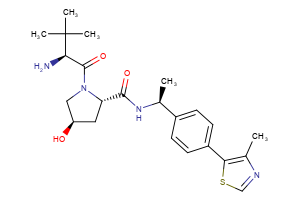
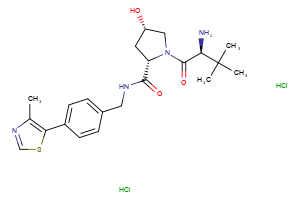
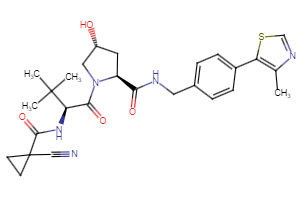
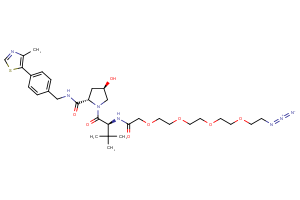
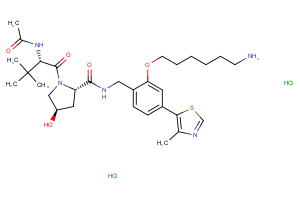
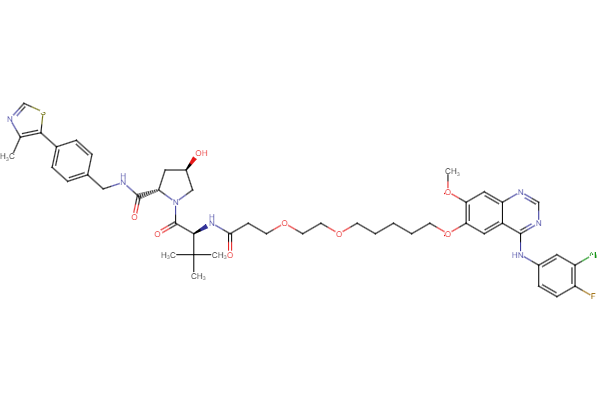
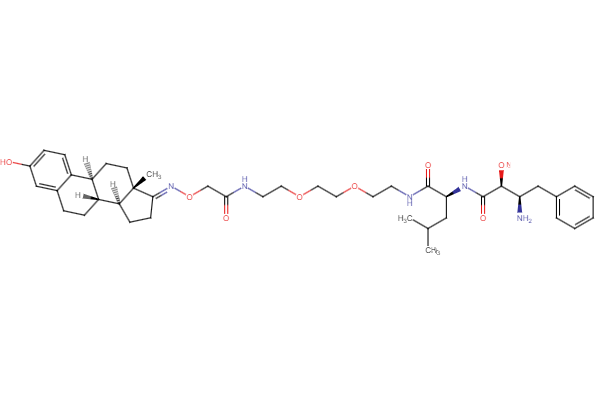
Selected fully functional TPD compounds, available from Key Organics
TPD Tool Molecules
Browse More Services
An overview of how Key Organics can be your solution provider.
Price competitive global leaders in fee-for-service synthetic organic chemistry provision.
Extensive experience in the development work required to take your project from gram to kilogram scale.
Carefully designed “three component” compound (E3 ligase binder, linker, protein-of-interest binder) to harness the cells endogenous waste disposal machinery.
More than 44,000 fragments that can be individually selected, allowing you to build bespoke fragment sets of size and composition best suited to your research needs.
All your key compound management activities including compound procurement, receipt, storage, formatting and distribution needs.
We offer a range of chemistry, sales and marketing, and technology consultancy services that can be specifically tailored to your needs.
High-end analytical services and support for the pharmaceutical, agrochemical, petrochemical and allied industries.